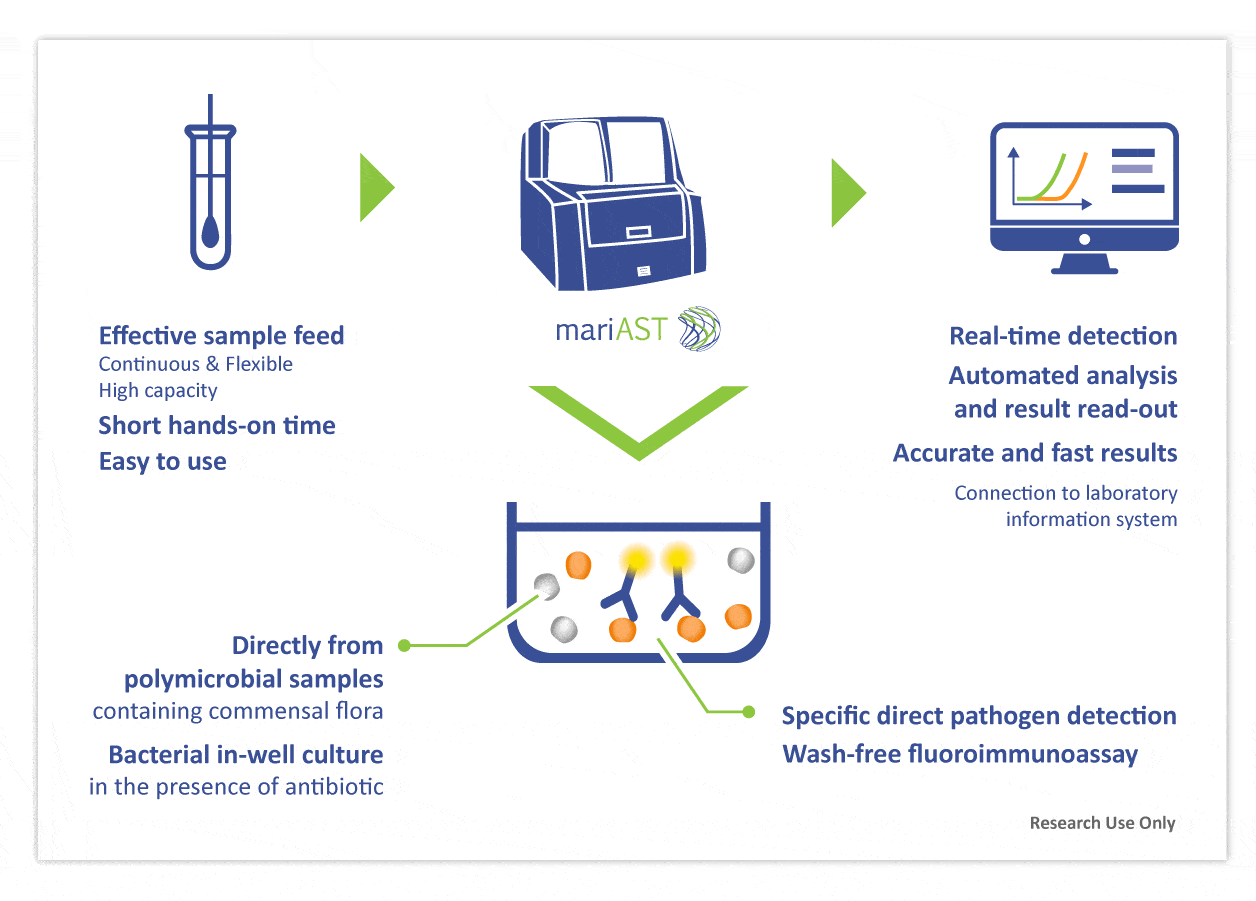
mariAST®- Rapid phenotypic AST without pure culture
The most advanced platform for rapid resistance testing
mariAST® combines in-well culture and specific bacterial detection. mariAST measures species-specific bacterial growth in real-time in the presence of an antibiotic.
The process yields to an identification in only 20 minutes for strong positive inocula. Phenotypic resistance results are reported in just hours after sampling and directly from samples containing commensal flora.
What is mariAST platform?
mariAST enables accurate resistance testing
- The first platform to allow phenotypic resistance testing directly from polymicrobial clinical samples containing commensal flora, and not just “directly from positive blood cultures”
- The only introduced platform capable of decentralized resistance testing and antibiotic susceptibility testing (AST)
- It is widely known that phenotypic resistance testing offers more reliable results than genotypic testing
- No compromise between assay sensitivity and result time
- Applies the same fully automated and field proven TPX technology as the CE marked mariPOC
mariAST results allow you to take the right decisions
- Pathogen- and strain-specific diagnosis and treatment
- Accurate use of antimicrobials
- Precise in-patient cohorting and fast discharge of contact isolation measures
- Improved cost-effectiveness and patient flow management
What applications mariAST is suitable for?
- The technology provides most added value when the time to result matters, the number of plausible pathogens is limited and testing breakpoint antimicrobial concentration is enough
- For example MRSA screening and testing, sexually transmitted diseases and non-penicillin susceptible pneumococci
- Technically mariAST enables also determination of MICs
mariAST is unique
A recent trend in microbiology is towards developing products for rapid phenotypic AST from positive blood culture bottles (sepsis). These methods, such as automated microscopy, are not suited for direct testing of samples containing commensal flora. Notably, sepsis AST, while of very high clinical importance, represents just a minor fraction of all AST applications.
Common infections are becoming untreatable due to the emergence of antimicrobial resistance (AMR). To diagnose and treat resistant infections, and to minimize emerging of resistance, the problem should be solved by decentralized rapid infection diagnostics combining both identification and resistance testing of pathogens. mariAST is the only presented platform technically and economically suitable for phenotypic and rapid AST in decentralized settings.
Antibiotic resistance
Today, antimicrobial susceptibility testing (AST) is a standard process used by clinical microbiology labs to analyse samples from hospitalized patients with infections. Currently, more than 700,000 people die of drug-resistant infections every year. The figure may reach 10 million by 2050. The emergence of new resistant bacteria and strains also means that there is an increasing need for antimicrobial susceptibility/resistance testing in outpatient wards in order to enable optimal treatment of patients suffering from common infections. So far these infections have been treated empirically.
Authorities and opinion leaders around the world have been calling for new technology solutions to tackle the problem of increasing antimicrobial resistance.
Reckless and incorrect use of antibiotics continues to increase the risk of spreading AMR. According to the World Health Organisation, AMR is the largest global health threat in the 21st century and it needs urgent measures. The focus of efforts to tackle the problem will be the decentralized rapid infection diagnostics combining both identification and resistance testing of pathogens.